Biochemical Sensors
Thanks to the biocompatibility of the employed materials, Organic Thin-Film Transistors (OTFTs) are commonly employed in biochemical sensing applications.
OTFTs can be roughly divides into Organic Field-Effect Transistors (OFETs) and Organic ElectroChemical Transistors (OECTs); both these structure can be employed ad biochemical sensors.
The DEALAB has a multi-year experience in the conception, development, fabrication and characterization of several kind of biochemical sensors, employing both OFETs and OECTs.
The contribution of the DEALAB to OFET-based biochemical sensors (chemFET and bioFET) is mainly represented by the employment of the CMFET working principle to biochemical sensing in liquid environment. In this field, the CMFET is characterized by several advantages with respect to other FET-based biochemical sensors, such as the possibility of being operated without the needing of an external, reference electrode in the measurement solution. Moreover, the organic CMFET has, as further advantage with respect to other OFET-based biochemical sensors, the physical separation between the bioreceptor layer and the organic semiconductor, which can be seriously damaged by the liquid measurement environment.
So far, OCMFET-based biochemical sensors has been so far employed in genetic analysis: as DNA is a negatively charged molecule, biochemical reactions related to an increase of its charge can be detected by the OCMFET and transduced in terms of threshold voltage shift.
Such working principle has been employed for DNA hybridization detection. DNA hybridization is that biochemical reaction which brings to the formation of the double-helix structure from two single-stranded, complementary DNA strands. Several genetic tests for medical, pharmaceutical and forensic applications are based on the detection of DNA hybridization. DNA hybridization detection is generally performed by indirect techniques, based on optical analysis, which requires costly equipment.
The OCMFET is able to perform a direct detection of DNA hybridization, by transducing its intrinsic charge. DNA sensitivity is obtained anchoring a single-stranded DNA molecule (probe) onto the sensing area; this operation, namely functionalization, brings to a first shift of the threshold voltage in the transistor structure, due to the negative charge of the probes. If the single-stranded DNA molecule complementary to the probe (target) is inserted on the sensing area, hybridization occurs and a further shift of the transistor’s threshold voltage is obtained, as a consequence of the increased negative charge anchored onto the sensing area.
So far, two different implementations of the OCMFET has been demonstrated to be able to detect DNA hybridization. A first, proof-of-concept device, employing a standard, high-voltage transistor was initially employed. As expected, a threshold voltage shift was obtained both after functionalization and hybridization; this last reaction was validated by means fluorescence tests.
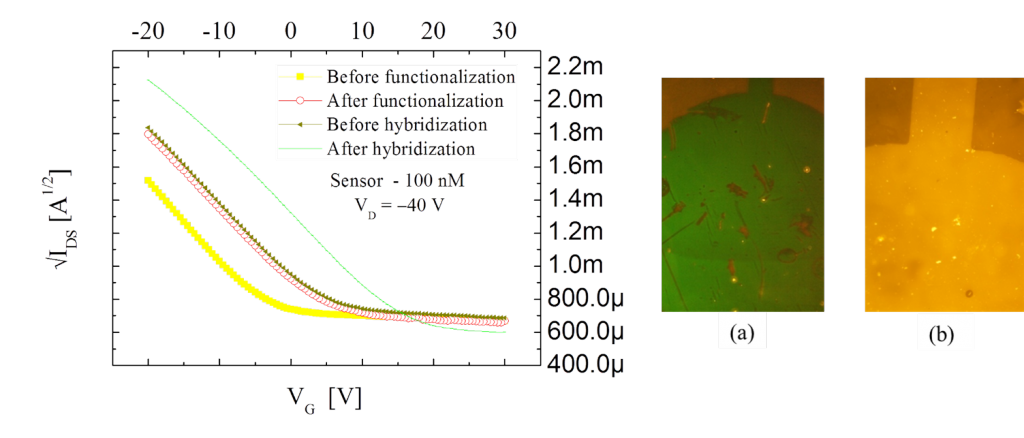
Figure 2: Shifts of the square-root of the transfer characteristic curves in the high-voltage, OCMFET-based DNA hybridization detection sensor after functionalization and hybridization processes; on the right, fluorescence picture of the sensing area before (a) and after (b) the functionalization and hybridization process with fluorescent dye-modified DNA sequences.
More recently, a novel implementation of the OCMFET has been developed. It is based on the low voltage OFET developed by our group, thus being able to be operated in the range of 2 V. The results confirm what obtained with the high voltage implementation (Figure 3(a)). Moreover, a complete calibration of the device was performed (Figure 3(b)), showing that the OCMFET is able to detect concentration as low as 100 pM; this result is the best ever reported in literature for OTFT-based DNA sensors. Finally, selectivity was investigated (Figure 3(c)). The results clearly show that a strong increase of the output current is obtained only if a fully-complementary target sequence is employed during hybridization; if only a base is changed in the sequence (single nucleotide polymorphism), a small increase of the current is obtained as a consequence of the weaker hybridization between probe and target sequences. On the contrary, the current decreased if a fully not-complementary target sequence was employed (i.e., hybridization does not occur).
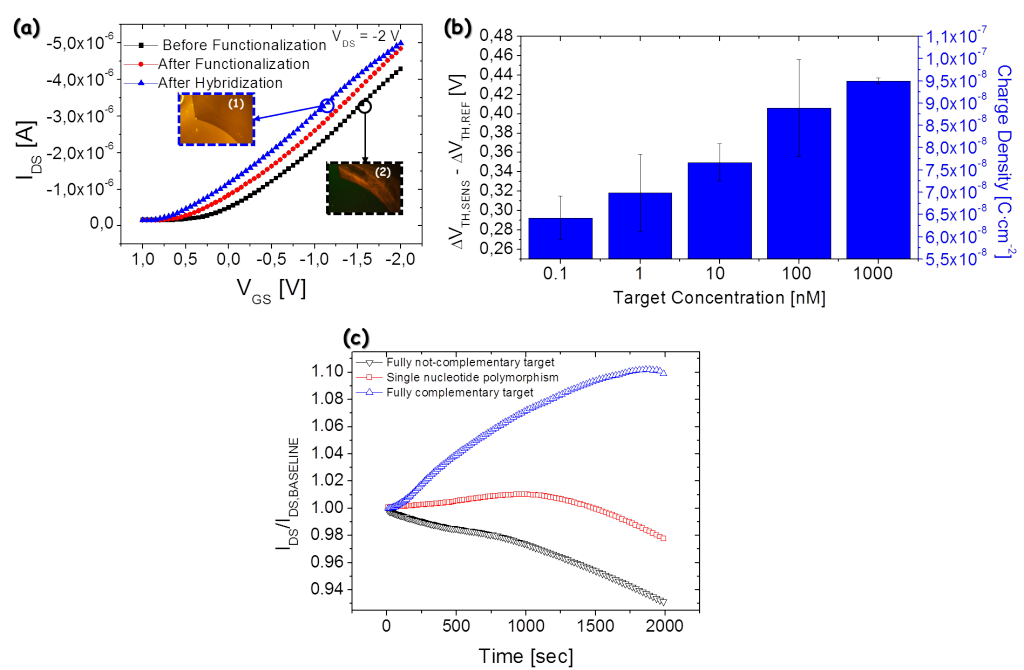
Figure 3: (a) shifts transfer characteristic curve during functionalization and hybridization processes; in the insets, fluorescent pictures of bare gold (1) and hybridized surface (2, with fluorescent dye-modified DNA target sequences) are reported; (b) calibration curve of the device with respect to target concentration in solution; (c) selectivity test, showing that a significant increase of the output current is obtained only if a fully complementary target sequence is employed in the hybridization process.
Related publications:
- M. Demelas, S. Lai, G. Casula, E. Scavetta, M. Barbaro, A. Bonfiglio, “An organic, charge-modulated field effect transistor for DNA detection”, Sensors and Actuators B: Chemical, vol. 171-172, pp. 198-203, 2012
- S. Lai, M. Demelas, G. Casula, P. Cosseddu, M. Barbaro, A. Bonfiglio, “Ultralow Voltage, OTFT‐Based Sensor for Label‐Free DNA Detection”, Advanced Materials vol. 25 (1), pp. 103-107, 2013
- M. Demelas, S. Lai, A. Spanu, S. Martinoia, P. Cosseddu, M. Barbaro, A. Bonfiglio, “Charge sensing by Organic Charge-Modulated Field Effect Transistors: application to the detection of bio-related effects”, J. Mater. Chem. B, 2013,1, 3811-3819
